Decoding Non-coding Genetic Reservoirs Shaping Life
In our research team, we address central questions in mammalian development and fecundity: How does transposable elements (TEs) function on cellular process and fate decision in host germline cells, and consequently impacting on developmental and reproductive fitness. About 15-20% of couples in industrialized country have difficulty conceiving a child. Although infertility is a serious clinical problem, basic knowledge of mechanisms ensuring integrity of gametes and embryonic development remains limited. Significantly, we and others have shown previously, that specific types of TEs exhibit distinctive spatiotemporal expression patterns during host gametogenesis and early stage of embryonic development. Furthermore, emerging evidence suggests that genetic and/or epigenetic ablations of the stage-specific expression of TEs contribute to various form of infertility and pregnancy loss.
TEs, mainly LINEs, SINEs and endogenous retroviruses (ERVs) that propagated within host genomes over generations, has account for a huge fraction of the modern mammalian genome, appearing in approximately 40-50% of the human and mouse genome. Note that TEs have dual nature; while they are traditionally considered selfish genetic elements and their mobilization can be detrimental to host genome integrity, specific types of TEs serves as the genetic reservoir to meet the environmental stress in a way that benefits the adaptive potential of host on an evolutionary scale. Indeed, evidence is mounting that integrated TEs into host genome −a.k.a., TEs co-option and domestication−, act as regulatory hubs modulating promoter-enhancer proximity, chromatin remodeling and 3D genome organization, which in turn impact on host cellular processes and multiple biological traits, as discovered in various species of flora and fauna.
As a premise, increase expression of TEs transcripts does not necessarily mean a proportional increase in (retro) transposition in host genome, because most of TEs have been inactivated by mutation and can no longer transpose within host genome. However, counterintuitively, specific types of TEs (particularly the young ERVs) are distinctively expressed during gametogenesis and early stage of embryonic development. Strikingly, our recent efforts demonstrated experimentally, that these spatiotemporal expressions of specific ERV subsets are essential during each step of reproductive life, including meiosis (to generate haploid gametes), onset of development after fertilization, and ontogeny. These finding are paradigm-shifting and will, for the first time, enable us to interrogate the functionalities of TEs throughout host germline development and to evaluate the impact of TEs-driven regulatory machinery on genome/epigenome stability, gene expression, fertility, and developmental plasticity across generations. Based on our exciting preliminary and published results, we propose a central hypothesis that the convergent function of co-opted TEs preserve genomic/epigenomic integrity of germline through two stratified mechanism ―(i) TEs-driven intricate cis-regulatory landscapes in regulating host gene expression and (ii) intimate relationship between TEs and host defense machinery in rewiring 3D genome structure and transcriptional network, initiating by recognition of non-self TEs sequences dispersed throughout the genome, thereby collectively serving as the functional genetic reservoirs (Figure 1). Furthermore, since TEs have invaded and expanded in genome at various points during biological evolution, suggesting that TEs-driven regulation of the host genome evolved in a species-specific manner (Figure 2). Our project contributes to not only closing the knowledge gaps in host-TEs dynamics and coevolution, but also providing new insight regarding the molecular functions of TEs during host germline development, which will be tremendous benefit to human and livestock reproductive medicine. We will address through the following specific AIMs to test this hypothesis.
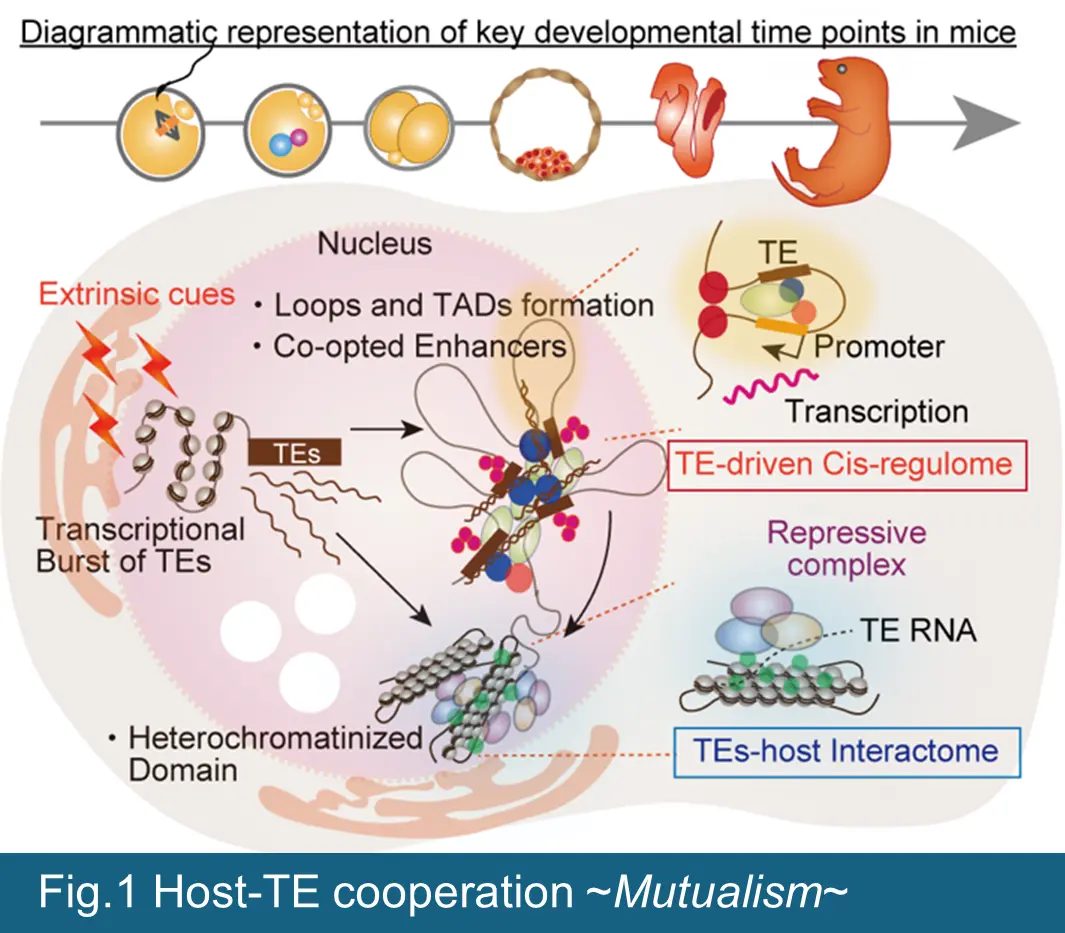
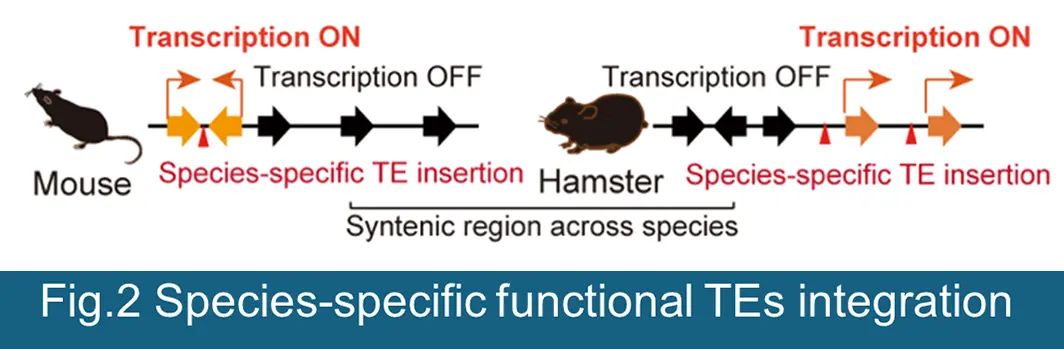
- AIM1Exploring the chromatin and transcriptional codes for the TEs during development & differentiation.
By leveraging recent advances in ultra-low input omics technologies, we will profile the epigenetic and transcriptional state of individual genomic copies of TEs during host development and cellular differentiation. Subsequently, through epigenetic and transcriptional network analyses and in silico simulation of host genes/TEs perturbation, we will explore new TEs candidates that are proposed to have a role in host developmental process. - AIM2Unraveling the molecular mechanism underlying the essential roles of TEs during development & differentiation.
We will use our existing targeting methodology for interspersed multiple TEs copies and the integrative approaches of ultra-low input HiC, ChIP-seq, and RNA-seq analyses to determine at the genome-wide scale how the loss-of-function of TEs affects chromatin structure and its remodeling during host development. - AIM3Deciphering the species-specific adaption of TEs in the regulation of host development.
We will explore species-specific diversity in TE-driven developmental programs through a cross-species comparative genomics, combined with above-mentioned omics approaches.
